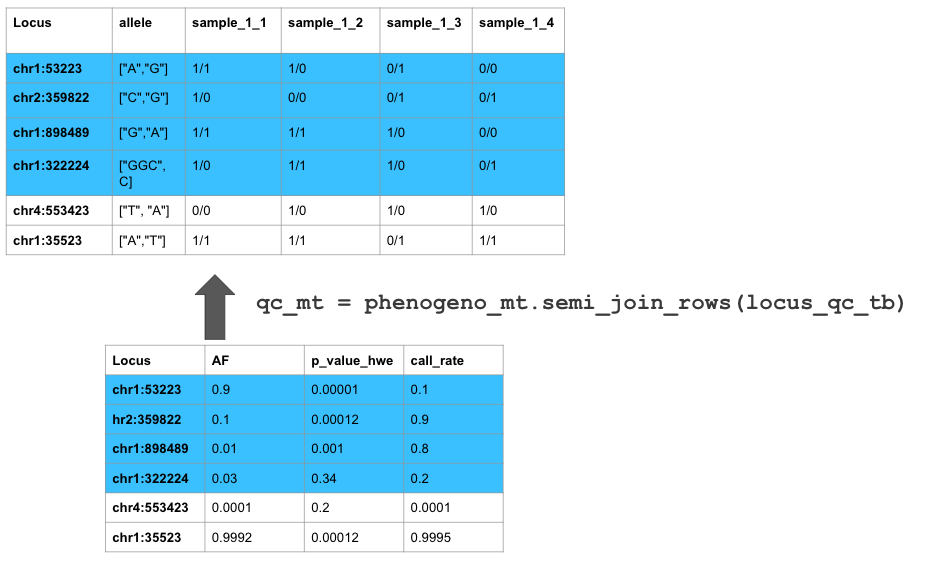
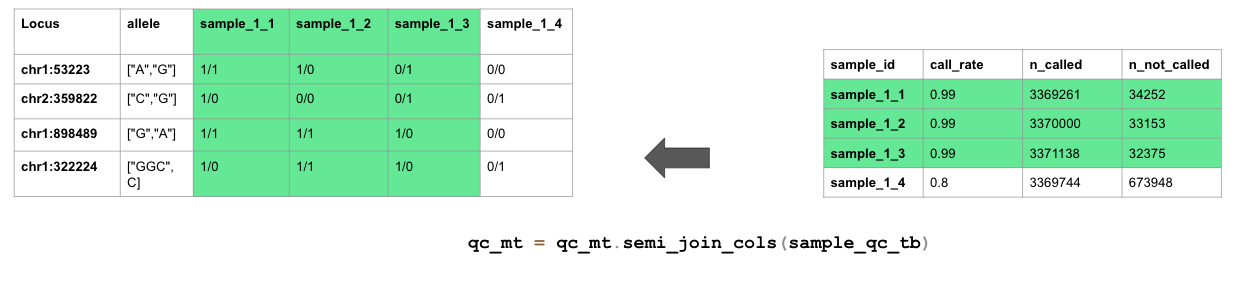
| chr1:12719 |
["G","C"] |
1.96e-02 |
1.35e-02 |
9.08e-01 |
3 |
True |
False |
[("ENSG00000223972.5","ENSG00000223972.5","DDX11L1","transcribed_unprocessed_pseudogene","2","gene",".","+",".")] |
"GRCh38" |
"G/C" |
NA |
[(NA,NA,NA,NA,"G/C",NA,NA,NA,12719,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,"rs1410641955",NA,NA,NA,NA,NA,NA,NA,12719,1)] |
NA |
12719 |
"." |
"chr1 12719 . G C . . GT" |
NA |
"splice_region_variant" |
NA |
NA |
"chr1" |
12719 |
1 |
[(1,NA,NA,"transcribed_unprocessed_pseudogene",NA,NA,NA,NA,NA,NA,NA,["intron_variant","non_coding_transcript_variant"],NA,NA,NA,"ENSG00000223972",NA,"DDX11L1","HGNC","HGNC:37102","ENST00000450305.2:n.182+22G>C",NA,NA,"MODIFIER","3/5",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,1,NA,"ENST00000450305",NA,NA,NA,"C"),(1,NA,NA,"processed_transcript",1,NA,466,466,NA,NA,NA,["splice_region_variant","non_coding_transcript_exon_variant"],NA,NA,"2/3","ENSG00000223972",NA,"DDX11L1","HGNC","HGNC:37102","ENST00000456328.2:n.466G>C",NA,NA,"LOW",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,1,NA,"ENST00000456328",NA,1,NA,"C"),(1,NA,NA,"unprocessed_pseudogene",1,NA,NA,NA,NA,NA,NA,["downstream_gene_variant"],1685,NA,NA,"ENSG00000227232",NA,"WASH7P","HGNC","HGNC:38034",NA,NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,-1,NA,"ENST00000488147",NA,NA,NA,"C"),(1,NA,NA,"miRNA",1,NA,NA,NA,NA,NA,NA,["downstream_gene_variant"],4650,NA,NA,"ENSG00000278267",NA,"MIR6859-1","HGNC","HGNC:50039",NA,NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,-1,NA,"ENST00000619216",NA,NA,NA,"C")] |
"SNV" |
0 |
0 |
| chr1:13380 |
["C","G"] |
-3.00e-01 |
9.08e-01 |
3.41e-01 |
5 |
True |
False |
[("ENSG00000223972.5","ENSG00000223972.5","DDX11L1","transcribed_unprocessed_pseudogene","2","gene",".","+",".")] |
"GRCh38" |
"C/G" |
NA |
[(NA,NA,NA,NA,"C/G",NA,NA,NA,13380,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,"rs571093408","G",8.20e-03,NA,NA,NA,NA,NA,13380,1)] |
NA |
13380 |
"." |
"chr1 13380 . C G . . GT" |
NA |
"splice_region_variant" |
[(1,["TF_binding_site_variant"],"N","MODIFIER",NA,"ENSM00080464946","ENSPFM0257",19,-2.10e-02,-1,"G")] |
[(1,"promoter_flanking_region",["regulatory_region_variant"],"MODIFIER",NA,"ENSR00001164745","G")] |
"chr1" |
13380 |
1 |
[(1,NA,NA,"transcribed_unprocessed_pseudogene",NA,NA,NA,NA,NA,NA,NA,["splice_region_variant","intron_variant","non_coding_transcript_variant"],NA,NA,NA,"ENSG00000223972",NA,"DDX11L1","HGNC","HGNC:37102","ENST00000450305.2:n.414+6C>G",NA,NA,"LOW","5/5",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,1,NA,"ENST00000450305",NA,NA,NA,"G"),(1,NA,NA,"processed_transcript",1,NA,628,628,NA,NA,NA,["non_coding_transcript_exon_variant"],NA,NA,"3/3","ENSG00000223972",NA,"DDX11L1","HGNC","HGNC:37102","ENST00000456328.2:n.628C>G",NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,1,NA,"ENST00000456328",NA,1,NA,"G"),(1,NA,NA,"unprocessed_pseudogene",1,NA,NA,NA,NA,NA,NA,["downstream_gene_variant"],1024,NA,NA,"ENSG00000227232",NA,"WASH7P","HGNC","HGNC:38034",NA,NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,-1,NA,"ENST00000488147",NA,NA,NA,"G"),(1,NA,NA,"miRNA",1,NA,NA,NA,NA,NA,NA,["downstream_gene_variant"],3989,NA,NA,"ENSG00000278267",NA,"MIR6859-1","HGNC","HGNC:50039",NA,NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,-1,NA,"ENST00000619216",NA,NA,NA,"G")] |
"SNV" |
0 |
0 |
| chr1:13494 |
["A","G"] |
-3.33e-01 |
6.83e-01 |
4.09e-01 |
5 |
True |
False |
[("ENSG00000223972.5","ENSG00000223972.5","DDX11L1","transcribed_unprocessed_pseudogene","2","gene",".","+",".")] |
"GRCh38" |
"A/G" |
NA |
[(NA,NA,NA,NA,"A/G",NA,NA,NA,13494,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,"rs574697788","G",1.40e-03,NA,NA,NA,NA,NA,13494,1)] |
NA |
13494 |
"." |
"chr1 13494 . A G . . GT" |
NA |
"non_coding_transcript_exon_variant" |
NA |
[(1,"CTCF_binding_site",["regulatory_region_variant"],"MODIFIER",NA,"ENSR00000344265","G"),(1,"promoter_flanking_region",["regulatory_region_variant"],"MODIFIER",NA,"ENSR00001164745","G")] |
"chr1" |
13494 |
1 |
[(1,NA,NA,"transcribed_unprocessed_pseudogene",NA,NA,456,456,NA,NA,NA,["non_coding_transcript_exon_variant"],NA,NA,"6/6","ENSG00000223972",NA,"DDX11L1","HGNC","HGNC:37102","ENST00000450305.2:n.456A>G",NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,1,NA,"ENST00000450305",NA,NA,NA,"G"),(1,NA,NA,"processed_transcript",1,NA,742,742,NA,NA,NA,["non_coding_transcript_exon_variant"],NA,NA,"3/3","ENSG00000223972",NA,"DDX11L1","HGNC","HGNC:37102","ENST00000456328.2:n.742A>G",NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,1,NA,"ENST00000456328",NA,1,NA,"G"),(1,NA,NA,"unprocessed_pseudogene",1,NA,NA,NA,NA,NA,NA,["downstream_gene_variant"],910,NA,NA,"ENSG00000227232",NA,"WASH7P","HGNC","HGNC:38034",NA,NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,-1,NA,"ENST00000488147",NA,NA,NA,"G"),(1,NA,NA,"miRNA",1,NA,NA,NA,NA,NA,NA,["downstream_gene_variant"],3875,NA,NA,"ENSG00000278267",NA,"MIR6859-1","HGNC","HGNC:50039",NA,NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,-1,NA,"ENST00000619216",NA,NA,NA,"G")] |
"SNV" |
0 |
0 |
| chr1:13504 |
["G","A"] |
1.50e-01 |
2.84e-01 |
5.94e-01 |
5 |
True |
False |
[("ENSG00000223972.5","ENSG00000223972.5","DDX11L1","transcribed_unprocessed_pseudogene","2","gene",".","+",".")] |
"GRCh38" |
"G/A" |
NA |
[(NA,NA,NA,NA,"G/A",NA,NA,NA,13504,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,"rs199896944",NA,NA,NA,NA,NA,NA,NA,13504,1)] |
NA |
13504 |
"." |
"chr1 13504 . G A . . GT" |
NA |
"non_coding_transcript_exon_variant" |
NA |
[(1,"CTCF_binding_site",["regulatory_region_variant"],"MODIFIER",NA,"ENSR00000344265","A"),(1,"promoter_flanking_region",["regulatory_region_variant"],"MODIFIER",NA,"ENSR00001164745","A")] |
"chr1" |
13504 |
1 |
[(1,NA,NA,"transcribed_unprocessed_pseudogene",NA,NA,466,466,NA,NA,NA,["non_coding_transcript_exon_variant"],NA,NA,"6/6","ENSG00000223972",NA,"DDX11L1","HGNC","HGNC:37102","ENST00000450305.2:n.466G>A",NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,1,NA,"ENST00000450305",NA,NA,NA,"A"),(1,NA,NA,"processed_transcript",1,NA,752,752,NA,NA,NA,["non_coding_transcript_exon_variant"],NA,NA,"3/3","ENSG00000223972",NA,"DDX11L1","HGNC","HGNC:37102","ENST00000456328.2:n.752G>A",NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,1,NA,"ENST00000456328",NA,1,NA,"A"),(1,NA,NA,"unprocessed_pseudogene",1,NA,NA,NA,NA,NA,NA,["downstream_gene_variant"],900,NA,NA,"ENSG00000227232",NA,"WASH7P","HGNC","HGNC:38034",NA,NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,-1,NA,"ENST00000488147",NA,NA,NA,"A"),(1,NA,NA,"miRNA",1,NA,NA,NA,NA,NA,NA,["downstream_gene_variant"],3865,NA,NA,"ENSG00000278267",NA,"MIR6859-1","HGNC","HGNC:50039",NA,NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,-1,NA,"ENST00000619216",NA,NA,NA,"A")] |
"SNV" |
0 |
0 |
| chr1:17358 |
["ACTT","A"] |
4.46e-01 |
1.30e+00 |
2.54e-01 |
5 |
True |
False |
[("ENSG00000227232.5","ENSG00000227232.5","WASH7P","unprocessed_pseudogene","2","gene",".","-",".")] |
"GRCh38" |
"CTT/-" |
NA |
[(NA,NA,NA,NA,"CTTCTTCT/CTTCT",NA,NA,NA,17366,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,"rs749387668",NA,NA,NA,NA,NA,NA,NA,17359,1)] |
NA |
17361 |
"." |
"chr1 17358 . ACTT A . . GT" |
NA |
"non_coding_transcript_exon_variant" |
NA |
NA |
"chr1" |
17359 |
1 |
[(1,NA,NA,"transcribed_unprocessed_pseudogene",NA,NA,NA,NA,NA,NA,NA,["downstream_gene_variant"],3689,NA,NA,"ENSG00000223972",NA,"DDX11L1","HGNC","HGNC:37102",NA,NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,1,NA,"ENST00000450305",NA,NA,NA,"-"),(1,NA,NA,"processed_transcript",1,NA,NA,NA,NA,NA,NA,["downstream_gene_variant"],2950,NA,NA,"ENSG00000223972",NA,"DDX11L1","HGNC","HGNC:37102",NA,NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,1,NA,"ENST00000456328",NA,1,NA,"-"),(1,NA,NA,"unprocessed_pseudogene",1,NA,582,584,NA,NA,NA,["non_coding_transcript_exon_variant"],NA,NA,"6/11","ENSG00000227232",NA,"WASH7P","HGNC","HGNC:38034","ENST00000488147.1:n.582_584del",NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,-1,NA,"ENST00000488147",NA,NA,NA,"-"),(1,NA,NA,"miRNA",1,NA,NA,NA,NA,NA,NA,["downstream_gene_variant"],8,NA,NA,"ENSG00000278267",NA,"MIR6859-1","HGNC","HGNC:50039",NA,NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,-1,NA,"ENST00000619216",NA,NA,NA,"-")] |
"deletion" |
0 |
0 |
| chr1:17373 |
["A","G"] |
-3.88e-02 |
4.94e-02 |
8.24e-01 |
4 |
True |
False |
[("ENSG00000278267.1","ENSG00000278267.1","MIR6859-1","miRNA","3","gene",".","-","."),("ENSG00000227232.5","ENSG00000227232.5","WASH7P","unprocessed_pseudogene","2","gene",".","-",".")] |
"GRCh38" |
"A/G" |
NA |
[(NA,NA,NA,NA,"A/G",NA,NA,NA,17373,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,"rs750111615",NA,NA,NA,NA,NA,NA,NA,17373,1)] |
NA |
17373 |
"." |
"chr1 17373 . A G . . GT" |
NA |
"splice_region_variant" |
NA |
NA |
"chr1" |
17373 |
1 |
[(1,NA,NA,"transcribed_unprocessed_pseudogene",NA,NA,NA,NA,NA,NA,NA,["downstream_gene_variant"],3703,NA,NA,"ENSG00000223972",NA,"DDX11L1","HGNC","HGNC:37102",NA,NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,1,NA,"ENST00000450305",NA,NA,NA,"G"),(1,NA,NA,"processed_transcript",1,NA,NA,NA,NA,NA,NA,["downstream_gene_variant"],2964,NA,NA,"ENSG00000223972",NA,"DDX11L1","HGNC","HGNC:37102",NA,NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,1,NA,"ENST00000456328",NA,1,NA,"G"),(1,NA,NA,"unprocessed_pseudogene",1,NA,NA,NA,NA,NA,NA,["splice_region_variant","intron_variant","non_coding_transcript_variant"],NA,NA,NA,"ENSG00000227232",NA,"WASH7P","HGNC","HGNC:38034","ENST00000488147.1:n.575-5T>C",NA,NA,"LOW","5/10",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,-1,NA,"ENST00000488147",NA,NA,NA,"G"),(1,NA,NA,"miRNA",1,NA,64,64,NA,NA,NA,["mature_miRNA_variant"],NA,NA,"1/1","ENSG00000278267",NA,"MIR6859-1","HGNC","HGNC:50039","ENST00000619216.1:n.64T>C",NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,-1,NA,"ENST00000619216",NA,NA,NA,"G")] |
"SNV" |
0 |
0 |
| chr1:17379 |
["G","A"] |
4.65e-01 |
2.92e+00 |
8.72e-02 |
4 |
True |
False |
[("ENSG00000278267.1","ENSG00000278267.1","MIR6859-1","miRNA","3","gene",".","-","."),("ENSG00000227232.5","ENSG00000227232.5","WASH7P","unprocessed_pseudogene","2","gene",".","-",".")] |
"GRCh38" |
"G/A" |
NA |
[(NA,NA,NA,NA,"G/A",NA,NA,NA,17379,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,"rs754322362",NA,NA,NA,NA,NA,NA,NA,17379,1)] |
NA |
17379 |
"." |
"chr1 17379 . G A . . GT" |
NA |
"mature_miRNA_variant" |
NA |
NA |
"chr1" |
17379 |
1 |
[(1,NA,NA,"transcribed_unprocessed_pseudogene",NA,NA,NA,NA,NA,NA,NA,["downstream_gene_variant"],3709,NA,NA,"ENSG00000223972",NA,"DDX11L1","HGNC","HGNC:37102",NA,NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,1,NA,"ENST00000450305",NA,NA,NA,"A"),(1,NA,NA,"processed_transcript",1,NA,NA,NA,NA,NA,NA,["downstream_gene_variant"],2970,NA,NA,"ENSG00000223972",NA,"DDX11L1","HGNC","HGNC:37102",NA,NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,1,NA,"ENST00000456328",NA,1,NA,"A"),(1,NA,NA,"unprocessed_pseudogene",1,NA,NA,NA,NA,NA,NA,["intron_variant","non_coding_transcript_variant"],NA,NA,NA,"ENSG00000227232",NA,"WASH7P","HGNC","HGNC:38034","ENST00000488147.1:n.575-11C>T",NA,NA,"MODIFIER","5/10",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,-1,NA,"ENST00000488147",NA,NA,NA,"A"),(1,NA,NA,"miRNA",1,NA,58,58,NA,NA,NA,["mature_miRNA_variant"],NA,NA,"1/1","ENSG00000278267",NA,"MIR6859-1","HGNC","HGNC:50039","ENST00000619216.1:n.58C>T",NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,-1,NA,"ENST00000619216",NA,NA,NA,"A")] |
"SNV" |
0 |
0 |
| chr1:17403 |
["A","G"] |
1.51e-01 |
1.41e-01 |
7.07e-01 |
5 |
True |
False |
[("ENSG00000278267.1","ENSG00000278267.1","MIR6859-1","miRNA","3","gene",".","-","."),("ENSG00000227232.5","ENSG00000227232.5","WASH7P","unprocessed_pseudogene","2","gene",".","-",".")] |
"GRCh38" |
"A/G" |
NA |
[(NA,NA,NA,NA,"A/G",NA,NA,NA,17403,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,"rs111588939",NA,NA,NA,NA,NA,NA,NA,17403,1)] |
NA |
17403 |
"." |
"chr1 17403 . A G . . GT" |
NA |
"non_coding_transcript_exon_variant" |
NA |
NA |
"chr1" |
17403 |
1 |
[(1,NA,NA,"transcribed_unprocessed_pseudogene",NA,NA,NA,NA,NA,NA,NA,["downstream_gene_variant"],3733,NA,NA,"ENSG00000223972",NA,"DDX11L1","HGNC","HGNC:37102",NA,NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,1,NA,"ENST00000450305",NA,NA,NA,"G"),(1,NA,NA,"processed_transcript",1,NA,NA,NA,NA,NA,NA,["downstream_gene_variant"],2994,NA,NA,"ENSG00000223972",NA,"DDX11L1","HGNC","HGNC:37102",NA,NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,1,NA,"ENST00000456328",NA,1,NA,"G"),(1,NA,NA,"unprocessed_pseudogene",1,NA,NA,NA,NA,NA,NA,["intron_variant","non_coding_transcript_variant"],NA,NA,NA,"ENSG00000227232",NA,"WASH7P","HGNC","HGNC:38034","ENST00000488147.1:n.575-35T>C",NA,NA,"MODIFIER","5/10",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,-1,NA,"ENST00000488147",NA,NA,NA,"G"),(1,NA,NA,"miRNA",1,NA,34,34,NA,NA,NA,["non_coding_transcript_exon_variant"],NA,NA,"1/1","ENSG00000278267",NA,"MIR6859-1","HGNC","HGNC:50039","ENST00000619216.1:n.34T>C",NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,-1,NA,"ENST00000619216",NA,NA,NA,"G")] |
"SNV" |
0 |
0 |
| chr1:17408 |
["C","G"] |
-8.03e-02 |
1.80e-01 |
6.71e-01 |
4 |
True |
False |
[("ENSG00000278267.1","ENSG00000278267.1","MIR6859-1","miRNA","3","gene",".","-","."),("ENSG00000227232.5","ENSG00000227232.5","WASH7P","unprocessed_pseudogene","2","gene",".","-",".")] |
"GRCh38" |
"C/G" |
NA |
[(NA,NA,NA,NA,"C/G",NA,NA,NA,17408,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,"rs747093451",NA,NA,NA,NA,NA,NA,NA,17408,1)] |
NA |
17408 |
"." |
"chr1 17408 . C G . . GT" |
NA |
"non_coding_transcript_exon_variant" |
NA |
NA |
"chr1" |
17408 |
1 |
[(1,NA,NA,"transcribed_unprocessed_pseudogene",NA,NA,NA,NA,NA,NA,NA,["downstream_gene_variant"],3738,NA,NA,"ENSG00000223972",NA,"DDX11L1","HGNC","HGNC:37102",NA,NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,1,NA,"ENST00000450305",NA,NA,NA,"G"),(1,NA,NA,"processed_transcript",1,NA,NA,NA,NA,NA,NA,["downstream_gene_variant"],2999,NA,NA,"ENSG00000223972",NA,"DDX11L1","HGNC","HGNC:37102",NA,NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,1,NA,"ENST00000456328",NA,1,NA,"G"),(1,NA,NA,"unprocessed_pseudogene",1,NA,NA,NA,NA,NA,NA,["intron_variant","non_coding_transcript_variant"],NA,NA,NA,"ENSG00000227232",NA,"WASH7P","HGNC","HGNC:38034","ENST00000488147.1:n.575-40G>C",NA,NA,"MODIFIER","5/10",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,-1,NA,"ENST00000488147",NA,NA,NA,"G"),(1,NA,NA,"miRNA",1,NA,29,29,NA,NA,NA,["non_coding_transcript_exon_variant"],NA,NA,"1/1","ENSG00000278267",NA,"MIR6859-1","HGNC","HGNC:50039","ENST00000619216.1:n.29G>C",NA,NA,"MODIFIER",NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,-1,NA,"ENST00000619216",NA,NA,NA,"G")] |
"SNV" |
0 |
0 |
| chr1:69735 |
["A","G"] |
7.29e-02 |
3.18e-02 |
8.59e-01 |
5 |
True |
False |
[("ENSG00000186092.6","ENSG00000186092.6","OR4F5","protein_coding","2","gene",".","+",".")] |
"GRCh38" |
"A/G" |
NA |
[(NA,NA,NA,NA,"A/G",NA,NA,NA,69735,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,NA,"rs1245478362",NA,NA,NA,NA,NA,NA,NA,69735,1)] |
NA |
69735 |
"." |
"chr1 69735 . A G . . GT" |
NA |
"synonymous_variant" |
NA |
NA |
"chr1" |
69735 |
1 |
[(1,"L","P2","protein_coding",NA,"CCDS30547.1",681,681,645,645,"ctA/ctG",["synonymous_variant"],NA,[("Gene3D","1"),("Pfam","PF13853"),("PROSITE_profiles","PS50262"),("PANTHER","PTHR26451"),("PANTHER","PTHR26451"),("Superfamily","SSF81321"),("Transmembrane_helices","TMhelix"),("CDD","cd15226")],"1/1","ENSG00000186092",NA,"OR4F5","HGNC","HGNC:14825","ENST00000335137.4:c.645A>G","ENSP00000334393.3:p.Leu215=",NA,"LOW",NA,NA,NA,NA,NA,NA,NA,NA,215,215,"ENSP00000334393",NA,NA,1,NA,"ENST00000335137",NA,NA,NA,"G"),(1,"L","A2","protein_coding",1,NA,768,768,708,708,"ctA/ctG",["synonymous_variant"],NA,[("Gene3D","1"),("PROSITE_profiles","PS50262"),("Transmembrane_helices","TMhelix"),("Superfamily","SSF81321"),("PANTHER","PTHR26451"),("PANTHER","PTHR26451"),("Pfam","PF13853"),("CDD","cd15226")],"3/3","ENSG00000186092",NA,"OR4F5","HGNC","HGNC:14825","ENST00000641515.2:c.708A>G","ENSP00000493376.2:p.Leu236=",NA,"LOW",NA,NA,NA,NA,NA,NA,NA,NA,236,236,"ENSP00000493376",NA,NA,1,NA,"ENST00000641515",NA,NA,NA,"G")] |
"SNV" |
0 |
0 |